I really thought I was done with the Express dplyr series though on completion of the second part I received many messages requesting more examples of using dplyr with ggplot along with some other types of information such as the Zika virus data which can be downloaded from Github. These examples are not drastically different from previous examples although they allow me to show how each data set presents its own challenges and also how transformations to the data will be necessary to visualize the information. The Zika data is spread across a number of countries but we will focus uniquely on cases in Brazil. Download the data using a Git client or straight from the site itself. If you are not using Git then you are missing out. (See my Youtube playlist for a basic introduction). It’s a great way to manage your R code and larger projects. Also keep in mind that the RStudio IDE has built in Git capabilities so as you create software it is extremely easy to “push” changes to your reference code and equally as easy for users of your code to “pull” changes down.
Processing the Data
 Okay we have seven .CSV files each of which represents some Zika information on the given date. (Note that there might be more files that have been added since I wrote this post). The format of the data isn’t particularly complicated although it isn’t standard in the sense that each row in a file represents the same type of information. In general you never really know what the data contains (or not) until you start working with it. There are a number of ways to approach this scenario. Ideally if there is a code book that explains the format then start there although it turns out that this data is simple enough to figure out. We could read in the data for a specific date, e.g. 2016-04-02.csv, but my inclination is to read in all the files and bind them together into a single data frame so I will have all that I need in one spot. This isn’t necessary but since the data itself is not very large then this is a reasonable approach.
Okay we have seven .CSV files each of which represents some Zika information on the given date. (Note that there might be more files that have been added since I wrote this post). The format of the data isn’t particularly complicated although it isn’t standard in the sense that each row in a file represents the same type of information. In general you never really know what the data contains (or not) until you start working with it. There are a number of ways to approach this scenario. Ideally if there is a code book that explains the format then start there although it turns out that this data is simple enough to figure out. We could read in the data for a specific date, e.g. 2016-04-02.csv, but my inclination is to read in all the files and bind them together into a single data frame so I will have all that I need in one spot. This isn’t necessary but since the data itself is not very large then this is a reasonable approach.
library(dplyr) # You will need to specify your own path to reflect your download location path <- "/Users/fender/Downloads/zika/Brazil/Epidemiological_Bulletin/data" setwd(path) temp_files <- list.files(pattern=".csv") temp_files [1] "Epidemiological_Bulletin-2016-04-02.csv" "Epidemiological_Bulletin-2016-04-23.csv" [3] "Epidemiological_Bulletin-2016-04-30.csv" "Epidemiological_Bulletin-2016-05-07.csv" [5] "Epidemiological_Bulletin-2016-05-14.csv" "Epidemiological_Bulletin-2016-05-21.csv" [7] "Epidemiological_Bulletin-2016-05-28.csv" # Next we read all seven files the contents of each will go into a list # element myfiles <- lapply(temp_files,read.csv,stringsAsFactors=FALSE) str(myfiles,1) List of 7 $ :'data.frame': 33 obs. of 9 variables: $ :'data.frame': 33 obs. of 9 variables: $ :'data.frame': 33 obs. of 9 variables: $ :'data.frame': 33 obs. of 9 variables: $ :'data.frame': 33 obs. of 9 variables: $ :'data.frame': 33 obs. of 9 variables: $ :'data.frame': 33 obs. of 9 variables:
The result of the list.files() function will be a character vector that contains the names of all the .CSV files in the current working folder. We then read all of these files in using the lapply() function. We could have used a for loop construct to do the same thing although the former approach is a more “R-like” way to do things. If you don’t yet understand lapply() or need a review of what it does then please see my posting which explains it in considerable detail. It is a very cool function in R that is used many places so you definitely want to get to get to know how it works. Each element of the resulting list contains a data frame that in turn contains the contents of one of the seven .CSV files. Let’s inspect the first two lines of the first two list elements to get an idea bout the Zika data.
# First let's get the column names for the data frames lapply(myfiles,names)[1] [[1]] [1] "report_date" "location" "location_type" "data_field" [5] "data_field_code" "time_period" "time_period_type" "value" [9] "unit" # Now look at the first two lines of the first two list elements lapply(myfiles,head,2)[1:2] [[1]] report_date location location_type data_field data_field_code time_period 1 2016-04-02 Norte region zika_reported BR0011 NA 2 2016-04-02 Brazil-Rondonia state zika_reported BR0011 NA time_period_type value unit 1 NA 6295 cases 2 NA 618 cases [[2]] report_date location location_type data_field data_field_code time_period 1 2016-04-23 Norte region zika_reported BR0011 NA 2 2016-04-23 Brazil-Acre state zika_reported BR0011 NA time_period_type value unit 1 NA 8545 cases 2 NA 716 cases</pre>
So we could work with each file/data frame individually although I want to create one large data structure. The do.call() function in R is used for a number of things but is frequently used to “bind” together individual list elements to form a single data structure so it’s going to be perfect for this job.
# The following will take all the individual list elements each of which are # individual data frames and bind them into a single data frame brazil <- do.call(rbind,myfiles) # Let's turn the report_date variable into a "real" date brazil <- brazil %>% mutate(report_date = as.Date(report_date)) glimpse(brazil) Observations: 231 Variables: 9 $ report_date <date> 2016-04-02, 2016-04-02, 2016-04-02, 2016-04-02, 2016-04-02,... $ location <chr> "Norte", "Brazil-Rondonia", "Brazil-Acre", "Brazil-Amazonas"... $ location_type <chr> "region", "state", "state", "state", "state", "state", "stat... $ data_field <chr> "zika_reported", "zika_reported", "zika_reported", "zika_rep... $ data_field_code <chr> "BR0011", "BR0011", "BR0011", "BR0011", "BR0011", "BR0011", ... $ time_period <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, ... $ time_period_type <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, ... $ value <int> 6295, 618, 375, 1520, 44, 771, 74, 2893, 30286, 1202, 7, 156... $ unit <chr> "cases", "cases", "cases", "cases", "cases", "cases", "cases... 9 NA 30286 cases
If we use the glimpse() function to examine this data frame we see that we have some basic variables represented such as report_date, location, and value which appears to represent the number of cases for a given location. The location_type variable tells us whether the location is a state or region with the latter being a container of possibly many states. The way I understand the information be organized is that on a certain date a .CSV file was uploaded that contained specific locations within Brazil and the number of reported Zika cases at that time. For each subsequent date (file) the same thing happened although the new number of cases are a “delta” or adjusted number to the previously reported number. We now have all of that in a single data frame. Let’s work with this data. We will remove columns 6 and 7 since all they have are missing values anyway as confirmed by the above glimpse() function results.
# Turn the data frame into a dplyr data table and get rid of # columns 5 and 7 brazil <- brazil %>% select(-(6:7)) # Let's look at the first twenty rows. While we are at it we will # omit columns 6 and 7 since the have all missing values anyway brazil %>% slice (1:20) # A tibble: 20 x 7 report_date location location_type data_field data_field_code value unit <date> <chr> <chr> <chr> <chr> <int> <chr> 1 2016-04-02 Norte region zika_reported BR0011 6295 cases 2 2016-04-02 Brazil-Rondonia state zika_reported BR0011 618 cases 3 2016-04-02 Brazil-Acre state zika_reported BR0011 375 cases 4 2016-04-02 Brazil-Amazonas state zika_reported BR0011 1520 cases 5 2016-04-02 Brazil-Roraima state zika_reported BR0011 44 cases 6 2016-04-02 Brazil-Para state zika_reported BR0011 771 cases 7 2016-04-02 Brazil-Amapa state zika_reported BR0011 74 cases 8 2016-04-02 Brazil-Tocantins state zika_reported BR0011 2893 cases 9 2016-04-02 Nordeste region zika_reported BR0011 30286 cases 10 2016-04-02 Brazil-Maranhao state zika_reported BR0011 1202 cases 11 2016-04-02 Brazil-Piaui state zika_reported BR0011 7 cases 12 2016-04-02 Brazil-Ceara state zika_reported BR0011 156 cases 13 2016-04-02 Brazil-Rio_Grande_do_Norte state zika_reported BR0011 640 cases 14 2016-04-02 Brazil-Paraiba state zika_reported BR0011 1060 cases 15 2016-04-02 Brazil-Pernambuco state zika_reported BR0011 333 cases 16 2016-04-02 Brazil-Alagoas state zika_reported BR0011 1479 cases 17 2016-04-02 Brazil-Sergipe state zika_reported BR0011 348 cases 18 2016-04-02 Brazil-Bahia state zika_reported BR0011 25061 cases 19 2016-04-02 Sudeste region zika_reported BR0011 35505 cases 20 2016-04-02 Brazil-Minas_Gerais state zika_reported BR0011 6693 cases
Understanding the Data
This data frame, though simple, is a little confusing. For each region a cumulative number of cases is presented which represents the sum of all the constituent states occurring in some number of rows below this record. For example – the first row of the data frame summarizes reported cases for the “Norte” region on 04/02/16 and then rows 2-8 are the constituent states and their respective case number totals. So the sum of the cases of rows 2-8 is the same as the case numbers reported on row 1. And then there are other entries for the “Norte” region for different reporting dates later on in the data table. If you look at row 19 you see that the same thing is happening for the “Sudeste” region. For someone wanting summary information this is fine but it makes the resulting data frame non-standard. As analysts you would probably want to have the region as a factor – another column in the data frame. Totals can easily be generated from this format so we lose nothing. But even with the current format we can still work with it. For example let’s pull out the totals for all regions and plot them.
# For each reporting_date we have a each of the 5 region case numbers
# So we have 7 dates/files so we have 35 rows in this table
brazil %>% filter(location_type=="region")
# A tibble: 35 x 7
report_date location location_type data_field data_field_code value unit
<date> <chr> <chr> <chr> <chr> <int> <chr>
1 2016-04-02 Norte region zika_reported BR0011 6295 cases
2 2016-04-02 Nordeste region zika_reported BR0011 30286 cases
3 2016-04-02 Sudeste region zika_reported BR0011 35505 cases
4 2016-04-02 Sul region zika_reported BR0011 1797 cases
5 2016-04-02 Centro-Oeste region zika_reported BR0011 17504 cases
6 2016-04-23 Norte region zika_reported BR0011 8545 cases
7 2016-04-23 Nordeste region zika_reported BR0011 43000 cases
8 2016-04-23 Sudeste region zika_reported BR0011 46318 cases
9 2016-04-23 Sul region zika_reported BR0011 2197 cases
10 2016-04-23 Centro-Oeste region zika_reported BR0011 20101 cases
# ... with 25 more rows
# Let's plot it
brazil %>% filter(location_type=="region") %>%
ggplot(aes(x=report_date, y=value,
group=location, color=location)) +
geom_line() +
geom_point() +
ggtitle("Brazil Zika Cases by Region")
Looks like the Southeast and Northeast have the most reported cases by far and the highest increase in reported cases over the available date period. This doesn’t necessarily mean that the actual number of cases in those regions changed that much over the given date range. It is possible that those cases were there all along and maybe the collecting and reporting mechanisms just caught up with the actual number of cases. (Note I have no idea if this is/was true or not). Let’s do a barchart of this information just for fun (and also to demonstrate an eccentricity of ggplot).
# Let's just pull out the regions into a table of its own
region <- brazil %>% filter(location_type=="region")
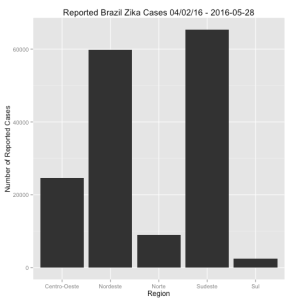
region %>%
ggplot(aes(x=location,y=value)) + geom_bar(stat="identity") +
ylab("Number of Reported Cases") + xlab("Region") +
ggtitle("Reported Brazil Zika Cases 04/02/16 - 2016-05-28")
Dealing with Factors
You saw something like this in the previous posting on dplyr. It’s a bar plot and it’s a bit unsatisfying since the bars are not in order according to number of cases. This data is easy to understand so it’s not a big deal although I don’t like this. I want it to be in order of most cases to least cases. To accomplish this will require creating a factor out of of the location variable based on the total number of reported cases. First here is the solution:
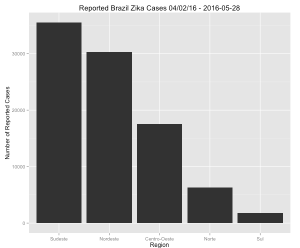
region %>%
slice(1:length(unique(region$location))) %>%
arrange(desc(value)) %>%
mutate(location=factor(location,levels=location,ordered=TRUE)) %>%
ggplot(aes(x=location,y=value)) + geom_bar(stat="identity") +
ylab("Number of Reported Cases") + xlab("Region") +
ggtitle("Reported Brazil Zika Cases 04/02/16 - 2016-05-28")
So how does this work ? Well check this out:
# This gets us the unique locations region %>% slice(1:length(unique(region$location))) report_date location location_type data_field data_field_code value unit 1 2016-04-02 Norte region zika_reported BR0011 6295 cases 2 2016-04-02 Nordeste region zika_reported BR0011 30286 cases 3 2016-04-02 Sudeste region zika_reported BR0011 35505 cases 4 2016-04-02 Sul region zika_reported BR0011 1797 cases 5 2016-04-02 Centro-Oeste region zika_reported BR0011 17504 cases # This arranges the unique locations by number of reported cases (descending) region %>% slice(1:length(unique(region$location))) %>% arrange(desc(value)) report_date location location_type data_field data_field_code value unit 1 2016-04-02 Sudeste region zika_reported BR0011 35505 cases 2 2016-04-02 Nordeste region zika_reported BR0011 30286 cases 3 2016-04-02 Centro-Oeste region zika_reported BR0011 17504 cases 4 2016-04-02 Norte region zika_reported BR0011 6295 cases 5 2016-04-02 Sul region zika_reported BR0011 1797 cases</pre> # So now we can create a factor out the locations variable using this order # Notice how location is now an ordered factor region %>% slice(1:length(unique(region$location))) %>% arrange(desc(value)) %>% mutate(location=factor(location,levels=location,ordered=TRUE)) %>% glimpse() Observations: 5 Variables: 7 $ report_date <date> 2016-04-02, 2016-04-02, 2016-04-02, 2016-04-02, 2016-04-02 $ location <ord> Sudeste, Nordeste, Centro-Oeste, Norte, Sul $ location_type <chr> "region", "region", "region", "region", "region" $ data_field <chr> "zika_reported", "zika_reported", "zika_reported", "zika_re... $ data_field_code <chr> "BR0011", "BR0011", "BR0011", "BR0011", "BR0011" $ value <int> 35505, 30286, 17504, 6295, 1797 $ unit <chr> "cases", "cases", "cases", "cases", "cases"
Standardizing the Data Format
We could continue to work with the data this way – with summaries for regions and the country sitting along side entries for states. But this mixture “violates” the standards of a “tidy” data frame. What I propose is to remove the summary info for the regions and country. We will make a factor column that gives us the region for each state. This is great because 1) all rows represent the same types of information and 2) we can easily recompute the totals for the region. There are a number of ways to do this – I’m just going with a quick method here with a loop.
# Here we will pull out these total so we can later check out work
brazil_totals <- brazil %>% filter(location=="Brazil")
region_totals <- brazil %>% filter(location_type=="region") %>%
group_by(report_date,location) %>%
summarize(tot=sum(value))
# Here we standardize the data frame and remove all summary rows because after all we can easily
# (re)compute any summary totals that we want
regvec <- vector() # A vector to hold the region for a given state
length(regvec) <- nrow(brazil)
for (ii in 1:nrow(brazil)) {
if (brazil[ii,]$location_type != "region") {
regvec[ii] <- newlab
} else {
newlab <- brazil[ii,]$location
regvec[ii] <- newlab
}
}
# Bind the region vector to the brazil data frame
statedf <- cbind(brazil,regvec)
# Next we eliminate the summary rows for the country and regions
statedf <- statedf %>% filter(location != "Brazil")
statedf <- statedf %>% filter(location_type != "region")
# Let's generate the region totals from the newly transformed data to make
# sure it matches what we had before.
statedf %>% group_by(report_date,regvec) %>%
summarize(tot=sum(value)) -> totals
# Do a spot check on the totals - compare this to the original data frame
# totals. There are differences in the column names but other than it's the
# same
all.equal(totals,region_totals)
[1] "Cols in y but not x: location. Cols in x but not y: regvec. "
# Let's take a sampling from this new data frame. Note how there is now
# a factor column that shows us what region a given state location belongs to
statedf %>% select(report_date,location,value,regvec) %>% sample_n(10)
report_date location value regvec
22 2016-04-02 Brazil-Santa_Catarina 62 Sul
174 2016-05-28 Brazil-Paraiba 2865 Nordeste
173 2016-05-28 Brazil-Rio_Grande_do_Norte 2312 Nordeste
140 2016-05-21 Brazil-Para 1583 Norte
48 2016-04-23 Brazil-Parana 1847 Sul
163 2016-05-28 Brazil-Rondonia 1032 Norte
101 2016-05-07 Brazil-Sao_Paulo 3452 Sudeste
136 2016-05-21 Brazil-Rondonia 974 Norte
67 2016-04-30 Brazil-Pernambuco 450 Nordeste
92 2016-05-07 Brazil-Rio_Grande_do_Norte 1757 Nordeste
# The following will give the same line plot as above just to show you that we
# can easily regenerate the totals with no problem.
statedf %>% group_by(report_date,regvec) %>% summarize(cases=sum(value)) %>%
ggplot(aes(x=report_date,y=cases,group=regvec,color=regvec)) +
geom_line() +
geom_point() +
ggtitle("Brazil Zika Cases by Region")
Drawing a Cluster Map
Okay what next ? Well how about generating a Leaflet Cluster Map where we see the number of cases reflected in a given region. And if we zoom in we can then see the sub clusters that make up the larger cluster. This is a cool feature of Leaflet. But to do this we will need to Geo Code the locations given in the data frame. This is easy to do since the ggmap package has geo coding capability built into it courtesy of the geocode() function which uses Google to do the work. See how easy this:
library(ggmap)
longlat <- geocode(unique(statedf$location)) %>%
mutate(loc=unique(statedf$location))
# Let's join this geocoded information to the statedf and save it into
# another data frame for mapping
statedf %>% filter(as.character(report_date)=="2016-05-28") %>%
group_by(location) %>% summarize(cases=sum(value)) %>%
inner_join(longlat,by=c("location"="loc")) %>%
mutate(LatLon=paste(lat,lon,sep=":")) -> formapping
head(formapping)
# A tibble: 6 x 5
location cases lon lat LatLon
<chr> <int> <dbl> <dbl> <chr>
1 Brazil-Acre 843 -70.00000 -9.00000 -9:-70
2 Brazil-Alagoas 3577 -36.00000 -9.00000 -9:-36
3 Brazil-Amapa 189 -52.00000 1.00000 1:-52
4 Brazil-Amazonas 2176 -63.00000 -5.00000 -5:-63
5 Brazil-Bahia 45419 -38.51083 -12.97111 -12.97111:-38.51083
6 Brazil-Ceara 2144 -38.54306 -3.71722 -3.71722:-38.54306
Wait. This doesn’t look right because we know we have many more cases in many more regions. What happened ? Well the way the cluster mapping option works is that it looks at each row and the corresponding latitude and longitude and clusters together similar rows. The data we mapped has the states listed once and their location so no wonder we get this limited output. If you look at the cases column it tells us how many times each cases occurred in that region. We need to find a hack to generate a data frame to get this number of cases for each location so the Leaflet Clustermap will look reasonable. There is a way to do this – a number of ways in fact. Here were will generate the row numbers of the data frame and for each row number we’ll replicate that row case number of times.
num_of_times_to_repeat <- formapping$cases
long_formapping <- formapping[rep(seq_len(nrow(formapping)),
num_of_times_to_repeat),]
head(long_formapping)
# A tibble: 6 x 5
location cases lon lat LatLon
<chr> <int> <dbl> <dbl> <chr>
1 Brazil-Acre 843 -70 -9 -9:-70
2 Brazil-Acre 843 -70 -9 -9:-70
3 Brazil-Acre 843 -70 -9 -9:-70
4 Brazil-Acre 843 -70 -9 -9:-70
5 Brazil-Acre 843 -70 -9 -9:-70
6 Brazil-Acre 843 -70 -9 -9:-70
# ah so this is the format we need
leaflet(long_formapping) %>% addTiles() %>%
addMarkers(clusterOptions=markerClusterOptions()
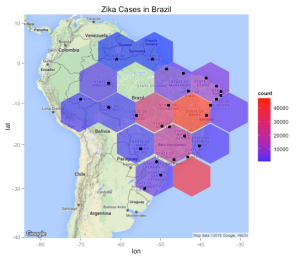
So this still isn’t ideal because if we click on one of the clusters to see the constituent sub clusters the performance can be slow especially in regions where there are lots of cases (like the Southest and Northeast). Why don’t we make a hexbin chart that might be more useful in representing the distribution of reported Zika cases in Brazil ? Hexbin charts are an extension of the heat map concept where squares are used to capture density or count in a region. The hexagonal shape is closer to a circle shape. What we will see is that the hex shapes on the map will be colored by count of Zika cases in that area. You will need to install the hexbin package prior to use. So it might be better to use a density geometry or a chloropleth map (see comments) though in creating the long_formapping data frame you will notice that it repeats the same latitude/longitude pair many times so in effect any resulting map is basically going to represent the counts of the same locales. We could jitter the lat/lon pairs some to get a better data set for density or chloropleth. For now we will stick with the hexbin since it overlays the map nicely.
library(hexbin) mymap <- qmap(c(lon=-56.09667,lat=-15.59611),zoom=4) mymap + geom_hex(bins=bins,data = long_formapping, aes(x = lon, y = lat),alpha=.6, inherit.aes=FALSE) + geom_point(data = long_formapping, aes(x = lon, y=lat), inherit.aes=FALSE,position = "jitter")+ scale_fill_gradient(low = "blue", high = "red")
As usual there are many other ways to kick this data round but I hope I have given you a realistic introduction to working with data that isn’t custom made for summary. Most of the data you will encounter will involve cleaning and manipulation prior to getting the best format for visualization and analysis. Speaking of analysis it can also be the case that you will need to juggle data formats around a few times to accommodate your interests. So many times you will never have a single ideal data format that will serve all your interests. This is why it is essential for you to become experienced with tools like dplyr, the lapply command, and some of the R programming constructs to glue it all together for you. Thanks for reading Steve Pittard © 2016 See my website for information on consulting and R training services.